DISCOVER
MOLECULAR
INTERACTIONS
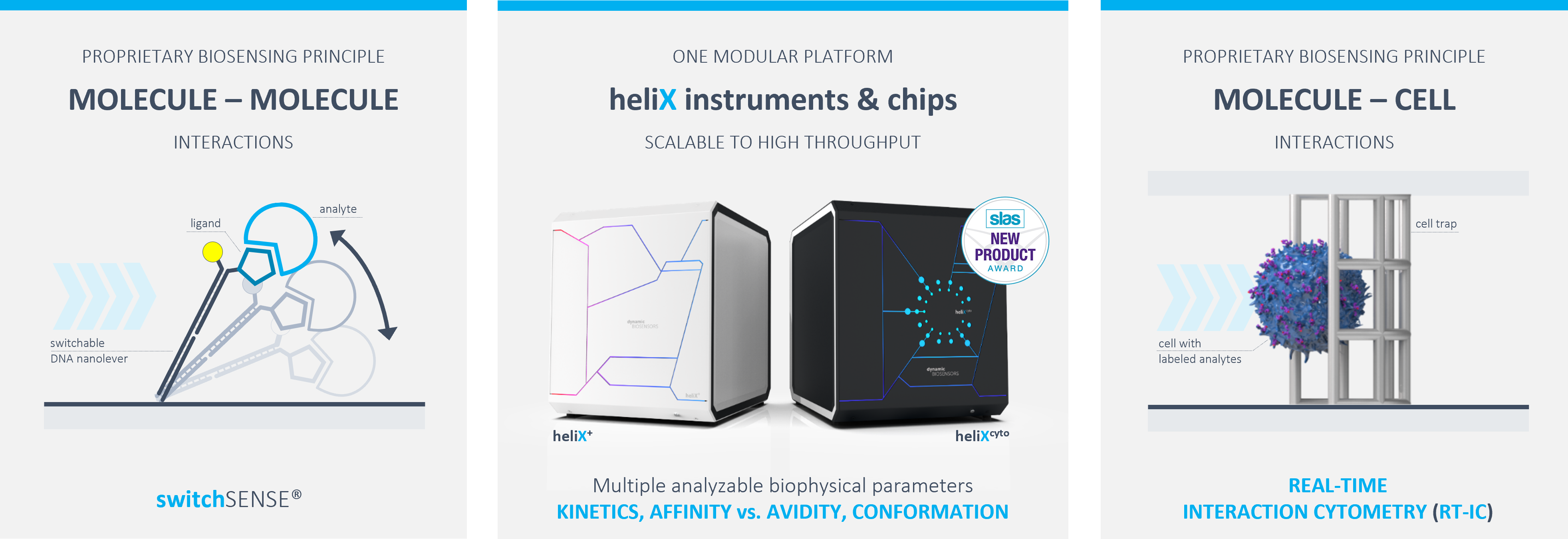
DISCOVER MOLECULAR INTERACTIONS using the heliX® platform – versatile assay formats for comprehensive biophysical information.
Every day, scientists all over the world strive to unravel the secrets of life. How do diseases develop? How can we treat them? In order to answer these big questions, we need to start small. We need to understand the tiny building blocks: molecules. How big are they? What is their structure? How do they interact with each other? The answers to these questions will guide us towards new and improved medicines to treat diseases like cancer.
At Dynamic Biosensors, we have made it our mission to support scientists in this endeavor and to help accelerate the drug discovery process.
To fulfil this, we developed switchSENSE® and Real-Time Interaction Cytometry (RT-IC) – two groundbreaking technologies that enable researchers to characterize molecular interactions in unequaled detail.
VIROLOGY
IMMUNOTHERAPY
ONCOLOGY
DRUG DISCOVERY & UNDRUGGABLE TARGETS
NEUROLOGICAL DISEASES
ENVIRONMENTAL TOXICOLOGY
RESEARCH TECHNOLOGY ADVANCES
BACTERIA
Our
Products
Made for high-performance analysis.
MOLECULE-MOLECULE INTERACTIONS
→ heliX / heliX+ biosensor instruments (switchSENSE® compatible)
MOLECULE-CELL INTERACTIONS
PURE PROTEIN-DNA CONJUGATES
„Until recently I always said, unfortunately we cannot do that on cells, kon and koff. A great answer now is with the heliXcyto we can. I like that a lot. Because this is really new and gives us new information.“
Prof. Harald Kolmar, Kolmar Group
TU DARMSTADT
In focus.
Browse through the latest publications.
New publication in antibodies I 2 May 2024 (open access)
Balancing the Affinity and Tumor Cell Binding of a Two-in-One Antibody Simultaneously Targeting EGFR and PD-L1
Congratulations to Julia Harwardt and our collaborators from Harald Kolmar‘s lab for their recent publication in antibodies!
The publication is about a specific type of symmetric bispecific antibodies called Two-in-One antibodies. The authors investigated whether affinity maturation for one target molecule (in this case EGFR) can be done without affecting the affinity for the second target molecule (in this case PD-L1). They compared mutated antibody variants with the wildtype antibody. They measured the binding kinetics of the antibodies to both targets simultaneously with switchSENSE®. Here, they made use of the technology’s unique capability to immobilize both target proteins on the surface at the same time in a controlled density and to perform dual-color measurements. This allowed them to investigate the influence of the two binding partners on each other. Moreover, they used RT-IC to measure the real-time binding kinetics of the antibody variants directly on double-positive tumor cell lines. They showed that especially the dissociation rate was reduced and thus antibody variants had a longer retention time on the cell surface.
New publication in the RSC Chemical Biology journal I 23 April 2024 (open access)
Reverse transcription as key step in RNA in vitro evolution with unnatural base pairs
Congratulations to Eva Hoffmann and our collaborators from Stephanie Kath-Schorr‘s lab for their insightful publication in RSC Chemical Biology!
They explored different reverse transcriptases and how they can be used to expand the genetic alphabet with unnatural bases. switchSENSE was used to monitor the binding affinities as well as the reverse transcription kinetics in real time and compare the kinetic efficacy of 2 different transcriptases on 4 different template ligand strands with 6 different natural and unnatural nucleotides. This study showcases that the heliX biosensor can be a powerful tool to study nucleic acid-modifying enzymes like reverse transcriptases and polymerases as it enables the real-time kinetic characterization of binding as well as activity.
New publication in eLife I 16 February 2024 (open access)
Repurposing the mammalian RNA-binding protein Musashi-1 as an allosteric translation repressor in bacteria
Congratulations to Roswitha Dolcemascolo and her co-authors for their publication on RNA-binding proteins in eLife!
The authors established the mammalian RNA-binding protein Musashi-1 (MSI-1) as a tool for synthetic biologists, specifically as a post-transcriptional repressor in E.coli.
switchSENSE® was used to study binding of this two-domain RNA binding protein to RNA ligands and to determine differences between the original RNA binding motif and mutated RNA ligands
New publication in the ACS Sensors journal I 7 December 2023 (open access)
Kinetic FRET Assay to Measure Binding-Induced Conformational Changes of Nucleic Acids
Congratulations to Anahi Higuera-Rodriguez and Mareike De Pascali for their recent publication in ACS Sensors!
They demonstrate the application of FRET in kinetic measurements for discerning binding-induced conformational changes in nucleic acids. This cutting-edge technique was applied to two distinct systems: the interaction between DNA and small molecules (MN19 aptamer with quinine) and the interaction between RNA and proteins (IMP3-KH1/2). The entire study was conducted on the heliX® platform.
The paper not only showcases the versatility and efficacy of FRET in unraveling intricate molecular dynamics but also sheds light on the specific interactions within the DNA-small molecule and RNA-protein systems studied. This work significantly contributes to our understanding of binding-induced conformational changes in nucleic acids.
NEWS.
News Feature | 20 March 2023
The Fraunhofer Institute for Cellular Therapies and Immunology (IZI-BB) has published a press release highlighting how switchSENSE® is used at the institute to characterize binding interactions of SARS-CoV-2.
The press release informs about the recent publication of Kruse et al. in Nature Scientific Reports, in which researchers investigated the interaction between the SARS-CoV-2 spike protein and its receptor hACE2. To do so, they prepared trivalent DNA-peptide nanostructures to achieve a trimeric hACE2 peptide presentation. switchSENSE® was used to examine the binding of full-length spike protein, subdomains and even whole virus particles. They show that small changes in the amino acid sequence of the spike protein can drastically influence the binding behavior. Furthermore, they demonstrate an increased affinity of the protein to the trivalent peptide construct as compared to single peptides. Finally, they detect different binding behaviors of pseudo virus models of the Alpha and Beta variants of SARS-CoV-2 compared to the inactivated wild type virus.
This study shows the versatility of the switchSENSE® technology, which enables the investigation of multivalent interactions and the utilization of a broad range of analytes from protein subdomains to whole virus particles.
This work was part of the project “CoronaSense” at the Fraunhofer Institute, which aims at using the switchSENSE® technology in order to characterize binding interactions of SARS-CoV-2 Spike proteins with its receptors in more detail to aid the development of anti-viral drugs but also of diagnostic tests.