switchSENSE®
eLearning Center
We cover an array of various, different applications.
Browse through publications, application notes,
tutorials, and more.
Molecule Type
Analytical Method

Insight into the avidity–affinity relationship of the bivalent, pH-dependent interaction between IgG and FcRn

Reverse transcription as key step in RNA in vitro evolution with unnatural base pairs

Real-time measurement of DNA polymerase activity and inhibition with switchSENSE® and the heliX® biosensor

Reverse transcription as key step in RNA in vitro evolution with unnatural base pairs

Repurposing the mammalian RNA-binding protein Musashi-1 as an allosteric translation repressor in bacteria

Balancing the Affinity and Tumor Cell Binding of a Two-in-One Antibody Simultaneously Targeting EGFR and PD-L1

DNA origami traps for large viruses
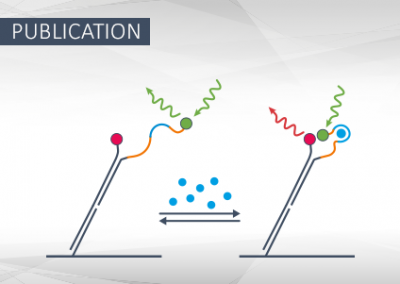
Kinetic FRET Assay to Measure Binding-Induced Conformational Changes of Nucleic Acids
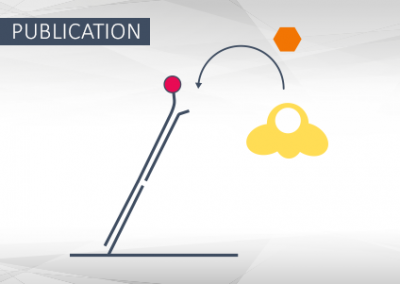
Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction

DISCOVER high throughput kinetic characterization of engineered multivalent peptide based inhibitors

DISCOVER protein conformational changes with electrically actuated DNA origami nanolevers

heliX Adapter Chip Storage and Handling

DISCOVER RNA protein interactions: Resolve multidomain interactions and RNA looping.
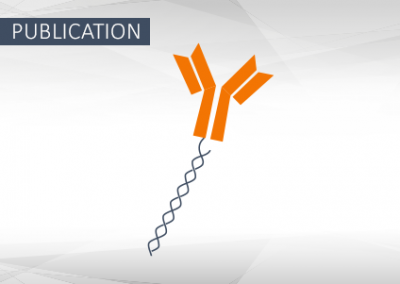
Programmable multispecific DNA-origami-based T-cell engagers
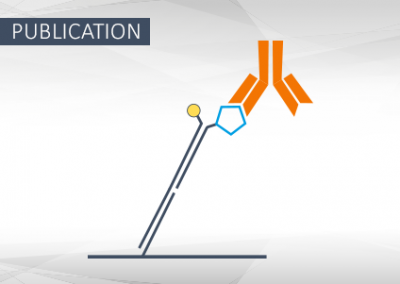
Synthetic Antigen-Conjugated DNA Systems for Antibody Detection and Characterization
DISCOVER complex membrane targets: measure binding to GPCRs, ion channels and more directly on cells
switchSENSE®: comprehensive characterization of molecule-molecule interactions

Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins

DISCOVER small molecule induced aptamer folding: a biosensor assay to detect structural changes

DISCOVER bispecific antibodies: straightforward characterization of mode of action & avidity effects

RT-IC Binding kinetics on cells
Kinetic analysis of ternary and binary binding modes of the bispecific antibody emicizumab

The Impact of Nε-Acryloyllysine Piperazides on the Conformational Dynamics of Transglutaminase 2
